PON01_02782 New nanotechnological strategies for the development of drugs and diagnostic tools directed to circulating cancer cells (CTC)
Leader
Biogem S.c.a r.l.
Duration
36 months
Keywords
CTC, antitumor, microalgae
ERC sectors
LS7_3, LS9_9
Summary of the project
The presence of circulating tumor cells (CTC) in the blood is associated with an advanced stage of the disease and reduce survival of cancer patients. Knowledge of the characteristics of the CTC is therefore of fundamental importance for many applications of clinical interest. This project is aimed to isolate CTC from the blood of patients suffering from epithelial and haematological tumors, to develop protocols for the ex-vivo expansion of these cells and to perform a complete molecular characterization in order to clearly define the mutational and transcriptional profiles. This project aims to develop diagnostic kits as well as to identify new molecular targets that may be the target of innovative therapeutic tools for the treatment of these tumors. The project also includes a phase which will identify molecules with anti-tumor activity against CTC, by screening libraries of compounds, substances and natural peptides including natural products arising from innovative sources such as marine microalgae.
Descriptions of activities
The project activities are divided into six objectives (OR).
O.R. 1 Development of nanotechnological devices for the isolation of circulating tumor cells (CTC)
OR2 Molecular characterization of CTC: identification of molecular targets
OR3 Molecular characterization of CTC: development of advanced diagnostic tools
OR4 Development of mouse models for the characterization of CTC
O.R. 5 Development of new technological procedures for targeting and CTC for the optimization of
delivery of drugs and / or molecules with antitumor activity
O.R. 6 Molecular Targeting of CTC
The Zoological Station is involved in the following activities:
Activity 6.2 Identification of molecules of natural and / or synthetic origin with possible biological activity on CTC cells.
The main objective of this activity is to provide extracts and fractions from marine cultivable microalgae with anti-tumor properties, and to isolate and characterize these compounds through a screening system based on biological assays of the fractions obtained from crude extracts. Finally we plan to optimize the production of bioactive compounds of interest through cultivation on a large scale of the producer microorganism(s).
Expected Results
Identification of at least one small molecule with antitumor activity on CTC cells.
Time chart

Tab 1 Costs
| Costs | Value | Duration |
|---|---|---|
| Total personnel costa | 200.000 | 36,00 |
| Total sub-contracting costs | 200.000 | 36,00 |
| Other costs | 160.000 | 36,00 |
| Equipments | 40.000 | 36,00 |
| General Costs | 100.000 | 36,00 |
| Total | 700.000 |
Tab 2 Personnel SZN
| MM/person | ||
|---|---|---|
| Ianora Adrianna | Senior researcher | 4 |
| Romano Giovanna | Researcher | 5,5 |
| Esposito Francesco | Technologist | 4 |
| Palumbo Flora | CTER | 7,6 |
| Perna Massimo | CTER | 7,6 |
| Biffali Elio | Technologist | 3 |
| Marino RIta | Technologist | 3 |
| Pannone Raimondo | CTER | 4 |
| Mauriello Elvira | CTER | 3 |
PON01_00117 Antigens and Adjuvants for Vaccines and Immunotherapy
Leader
Novartis Vaccines & Diagnostics Ltd.
Duration
36 months
Keywords
Biotechnology, human health, microalgae
ERC sectors
LS7_3, LS6_11, LS7_9
Summary of the project
The therapeutic benefits of a stimulation of the innate immune system are well documented. Many substances of natural origin, including some lipidic or glycolipidic molecules are able to stimulate various cellular components of the innate immune system. Therefore metabolites of low molecular weight or lipid components of extracts of marine eukaryotic microorganisms may have activating properties of the innate immune cells and act as adjuvants or immune-modulators.
The innovative approach proposed in this project is based on a public-private partnership and belongs mainly to Areas of Convergence (Sicily, Calabria and Campania).
The project aims to achieve three important goals: 1. The development of innovative vaccines for bacterial and viral infections 2. The development of new molecules with adjuvant action and the study of the mechanism of action of those already known 3. The development of more effective and safe new viral vectors for the development of new vaccines.
The role of the Zoological Station in the project is to identify and cultivate species of marine microalgae that show potential antigen and adjuvant activities.
Descriptinos of the activity
The project involves 3 objectives:
OR 1. Identification of antigenic candidates for the development of potential vaccines against Streptococcus pneumoniae, Clostridium difficile, Escherichia coli pathogens, HCMV
OR 2. Identification of new adjuvants and analysis of the mechanism of action of known adjuvants
OR 3. Identification and validation of new Adenovirus and MVA vectors and Virus Like Particles for use as potential vaccines against infectious diseases (HCMV, Malaria, Influenza, RSV, etc.)
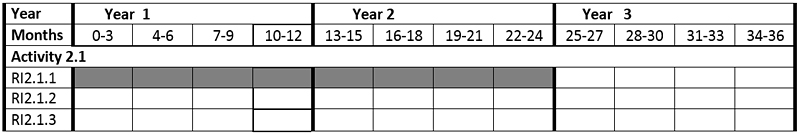
The Zoological Station participates in activity 2.1
Activity 2.1. Identification of novel natural compounds with immune-modulatory and adjuvant activity from marine microalgae
and in particular in:
RI 2.1.1. Preparation of extracts and fractions from microalgae, isolation and identification of substances with immune-regulatory properties (Stazione Zoologica).
Extracts of marine organisms have already been shown to contain immune-regulatory substances (eg alpha galoctoside ceramide that can stimulate NKT cells) and lipids other than those present in humans. This project aims not only to identify new molecules able to interact with the immune system, but also to identify compounds for therapeutic formulations, for example compounds as adjuvants for vaccines. To this end, the group of dr. Ianora is preparing pellets of microalgae for the isolation and identification of substances with potentially immune-regulatory activity. The fractions are assayed by industrial partners (Novartis) for their ability to activate various cells and receptors of the innate immune system. The study will be performed in collaboration with IBB-CNR (SZN sub-contractor) who will characterize the various active components through the use of NMR techniques.
Time chart
Expected results
To obtain at least one new natural compound from marine microalgae with immuno-stimulatory or adjuvant activity.
Tab 1 Costs
| Costs | Value | Total duration (months) |
|---|---|---|
| Total personnel costs | 435.000 | 24 |
| Total sub-contracting costs | 140.000 | 24 |
| Other costs | 318.500 | 24 |
| General costs | 217.500 |
Tab 2 Personnel SZN
| Total MM/person | ||
|---|---|---|
| Ianora Adrianna | Senior researcher | 8 |
| Romano Giovanna | Researcher | 8 |
| Esposito Francesco | Technologist | 10 |
| Palumbo Flora | CTER | 10 |
| Perna Massimo | CTER | 10 |
| Cter 2 temp determinato | CTER | 22 |

Project financed in the framework of PON R&C 2007-2013 - PAC Enhancement of public research infrastructures
Coordinator Raffaella Casotti
The project concerns the enhancement of research infrastructures for marine environment located in the Convergence Regions Sicily, Campania and Puglia, where the sea is a primary opportunity of development. The project, called EMSO‐MedIT is the Italian contribution to the consolidation in the above mentioned regions of the European research infrastructure EMSO, which, within the context
of EMSO‐MedIT, is in synergy with the other ESFRI coordinated by Italy (KM3NeT and EMBRC) and the Italian initiative for marine research RITMARE.
The actions foreseen will be carried out according to the following objectives:
1) enhancement of marine infrastructures and of scientific and technological facilities to strengthenand expand the network for multidisciplinary monitoring of coastal, deep and the water column
marine environment;
2) networking of all existing and enhanced infrastructures for the transmission in real‐time/near‐realtimeintegrating the data from fixed and relocatable observing systems;
3) establishment of a mobile system of intervention to be used for monitoring campaigns at sites ofstrategic interest or in the case of environmental emergencies.
The network of monitoring infrastructures will be further enhanced through the creation of anexchange information system that will enable the sharing of the large amounts of data, providing access to a large community of Italian and foreign users.
Our role: We are partners of the project and in charge of the WP2 "Strengthening of Campania", together with INGV for the Gulf of Pozzuoli. The expansion includes the acquisition of different oceanographic instrumentation, including a WaveGlider, an ROV, and various sensors, but mainly two buoys type elastic beacon to locate in the Gulf of Naples and Gulf of Pozzuoli for the real-time monitoring and data transmission to the control center of physical and biological environmental data.
Partners: National Institute of Geophysics and Volcanology (INGV), Anton Dohrn Zoological Station (SZN), National Institute of Nuclear Physics (INFN), the National Research Center (CNR), National Institute for Environmental Protection and Research (ISPRA)
PON01_02093 Study of new technologies and technological platforms for the improvement of production processes of active pharmaceutical ingredients of industrial interest and search for new bioactive molecules from natural sources
Leader
Sanofi-Aventis S.p.A.
Duration
36 months
Keywords
natural medicines, chemotherapy, microalgae
ERC sectors
LS7_3, LS9_9
Summary of the project
The overall objective of the project is the study and application of advanced and innovative technologies for the improvement of productive processes of pharmaceutical industry and the search for new molecules with potential pharmacological activity in the anti-infective, anti- cancer and anti-inflammatory field.
The first research line of the project will study the most innovative aspects of technologies of microbiology and genetics of producer strains.
The second research line will study the possibility to identify new products as candidates of potential pharmaceutical interest in the fields of anti-infectives and more generally in cancer and chronic-degenerative diseases connected with ageing, with particular attention to inflammation role. These activities will be focused on the search for new substances of pharmacological interest by screening extracts from microorganisms and / or aquatic organisms and on the characterization of their beneficial and anti-infection properties.
The project will allow the maintenance of a high scientific and technological concentration of great innovative potential characterized by an organic collaboration between industrial and academic researchers.
Description of activities
The project is divided into two lines, each divided into four development objectives (OR) according to the following scheme:
OR 1.1 - Genetic Improvement / genomic technologies
OR 1.2 - Genetic Improvement / Strain improvement
OR 1.3 - Physiology of fermentation
OR 1.4 - Extraction / Purification
OR 2.1 - New methods for the search of bioactive molecules by microorganisms
OR 2.2 - Summary of chemical derivatives of mature products
OR 2.3 - Identification of target and natural compounds relevant to cancer diseases, and chronic degenerative diseases associated with aging
OR 2.4 - Screening and characterization of extracts from marine organisms
The Zoological Station will be involved in the following activities:
OR 2.4 - Screening and characterization of extracts from marine organisms
The purpose of this Objective is the identification of new active principles with antimicrobial and / or antitumor and / or protective activity against neurodegeneration and / or aging. To achieve this goal the Zoological Station will perform the following activities:
ARI 2.4.1 - Identification of bodies from which to extract the active ingredients and their collection
Will be identified already known that microalgae can be a source of active ingredients with antimicrobial activity, antitumor, antineurodegenerativa, anti-aging. The selection will be made on the basis of their environmental activities.
ARI 2.4.2 - Extraction and sample preparation
Each species of microalgae will be cultivated to SZN so as to obtain a sufficient biomass for the extraction of small molecules by the Institute of CNR of Biomolecular Chemistry (ICB-CNR) as a third party custodial .. Once you have identified the active fractions from the other partners will identify molecule chemistry by the ICB and production in the SZN algae that produce these molecules.
Expected Results
Identification of at least one product that has the characteristics to be evaluated as a new "lead candidate" (New Product).
Time charts
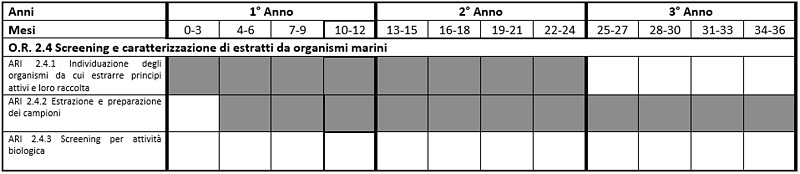
Table 1 Costs
| Costi | Valore | Durata mesi |
|---|---|---|
| Spese di personale totale | 360.000 | 36 |
| Costi consulenza tot | 225.000 | 36 |
| Altri costi esercizio totali | 160.000 | 36 |
| Spese generali | 180.000 | 36 |
Table 2 Personnel SZN
| Nominativo | Qualifica | Monte ore tot per persona | Durata mesi totale |
|---|---|---|---|
| Ianora Adrianna | Senior Researcher | 5,5 | 36 |
| Romano Giovanna | Researcher | 8 | 36 |
| Esposito Francesco | Technologist | 8 | 36 |
| Palumbo Flora | Cter | 8 | 36 |
| Perna Massimo | Cter | 8 | 36 |
| Tecn 1 temp det | Technologist | 22 | 24 |
| Cter 1 tempo det | Cter | 22 | 24 |
| Totale ore | 82 | 36 |
IRMA: Implementation and Remote Connection for Real Time Moniotoring of Marine Microorganisms
Funded by the Italian Ministry of University and Education (MIUR)
Scientific Coordinator Raffaella Casotti
This project proposes to use a prototype for the automatic, continuous staining of bacteria for the in situ high frequency monitoring (several times a day, up to every 30 min). This instrument will complement the tools already available to SZN for the monitoring of photosynthetic microorganisms, allowing the definition in close temporal and spatial scale of the microbial compartment. We will study the feasibility of use on sampling boats, even without any technical supervision and, in the long term, on buoys. The data produced will provide useful information on the microbial dynamics at very small time scale in several coastal sites. The final product will be a demonstrator of multiparameter monitoring station that transmits environmental data from different sources in real time, operated remotely, helping to build an early warning system for environmental risk
Our role in the project is to coordinate actions and to test, calibrate and validate the prototype
Partners: SZN, CytoBuoy bv (Holland, George Dubelaar), Eawag (Switzerland, Frederick Hammes), CNRS- MIO (France, Gérald Grégori, Melilotus Thyssen, Michel Denis), INGV (Italian, Giovanni Iannaccone, Sergio Guardato)