Tenders and Job Opportunities
Tenders and Job Opportunities Job calls
Job calls Scholarships and research fellowships
Scholarships and research fellowships Open Calls
Open Calls ENGLISH
ENGLISH Pages
Pages
Pages
PharmaDeep
Summary
This project aims to collect samples at the South Shetland Trough (SST) near the Antarctic Peninsula, one of the few cryogenic deep-sea trenches in the world.
The specific aims of PharmaDeep are to:
(a) Collect marine organisms from deep-and-cold-water habitats that may be unique sources of natural products for the treatment of cancer and infectious diseases.
(b) Perform the first marine biological survey of the unique habitat, the SST.
(c) Compare and contrast the SST fauna and drivers of distribution with other trench ecosystems.
(d) Investigate the interaction of physical and chemical extremes of relevance to understanding the habitability of other planetary bodies.
What we do
SZN will be mainly involved in Zooplankton and Phytoplankton sample collection for taxonomy identification, activity screening investigation and later chemical analysis.
Partners
SZN, IBP-CNR, University of Aberdeen, Universitat de Barcelona, Insituto Español de Oceanografía, BioBridge, University of Chile, Nicolaus Copernicus University, Edinburgh University
Research Area
Marine Biotechnology
SZN role
Partner
Principal Investigator
Project Manager
Alan Jamieson
Project Lifetime
October 2015 - June 2016
Funding Institutions
European Commission, under the EU EUROFLEETS2 program grant agreement n° 312762
Personnel involved
Adrianna Ianora, Principal investigator
Chiara Lauritano, Post-doc
Christian Galasso, PhD Student
PharmaSea

Summary
The PharmaSea project focuses on marine biodiscovery research, development and commercialization and brings together a broad interdisciplinary team of academic and industry researchers and specialists to produce new products for development in three accessible market sectors, health (infection, inflammation, CNS diseases), personal care and nutrition. Despite the enormous potential of blue biotechnology, the exploitation of marine natural products, in particular at the commercial scale, has been hindered by a number of constraints. These concern the access of organisms (physical and legal), their genetics, bioactive compound isolation, elucidation of chemical structures and reliable early validation of the biological activity of these compounds. PharmaSea directly aims to overcome some of these bottlenecks leading to improvements in the quality of marine resources available for biotechnological exploitation, to shorten time to market, and develop sustainable modes of supply of raw materials for industry. Within the project a wide variety of marine microorganisms will be analyzed, including collections held by some partners and new collections of strains collected in extreme environmental conditions (deep, hot and cold) in order to isolate new compounds with characteristics appropriate for development by SMEs. The overall objective of PharmaSea is to produce two compounds at large-scale and promote them in pre-clinic evaluations.
What we do
SZN is involved in WP1 and WP2 of this project. SZN objectives are to select and isolate microalgae with potential pharmaceutical activity (e.g. antimitotic, anti-bacterial or for neurodegenerative diseases), and to develop innovative technologies for cell growth in order to increase the production of specific secondary metabolites. Often canonical growth conditions do not allow the activation of specific metabolic pathways. Therefore it is necessary to experiment with new growth conditions in order to obtain a greater concentration of the desired product. Moreover, the aim of the SZN is, also, to scale-up the production of selected strains using 100L photobioreactors under controlled conditions. Molecular techniques are also being used to identify species and to study the transcriptomes of microalgal strains that show biological activity.
Partners
SZN, Katholieke Universiteit Leuven, The University Court of the University of Aberdeen, MarBio, eCoast, Biodridge, MEDINA, University college Cork, BIOCOM, CNR, Universidade de Santiago de Compostela, The Royal society of chemistry, C-LECTA GMBH, Denmarks Tekniske Universitet, DEEPTEK, Advanced chemistry development UK, Wuhan University, Institute of Microbiology Chinese Academy of Sciences, University of the Western Cape, Instituto de Dinamica Celular y Biotecnologia, Asociacion Instituto Nacional de Biodiversidad, Union Internationale pour la conservation de la nature et de ses ressources, University of Waikato, SeaLifePharma.
Research Area
Marine Biotechnology
SZN role
Partner
Principal Investigator
Project Manager
Peter de Witte
Project Lifetime
October 2012 - October 2016
Funding Institutions
European Commission, under the 7th Framework Programme (FP7/2007-2013 under grant agreement n° 312184)
Personnel involved
Adrianna Ianora, Principal investigator
Giovanna Romano, Experienced Researcher
Francesco Esposito, Technologist
Chiara Lauritano, Post-doc
PON 02782 - Biogem
PON01_02782 New nanotechnological strategies for the development of drugs and diagnostic tools directed to circulating cancer cells (CTC)
Leader
Biogem S.c.a r.l.
Duration
36 months
Keywords
CTC, antitumor, microalgae
ERC sectors
LS7_3, LS9_9
Summary of the project
The presence of circulating tumor cells (CTC) in the blood is associated with an advanced stage of the disease and reduce survival of cancer patients. Knowledge of the characteristics of the CTC is therefore of fundamental importance for many applications of clinical interest. This project is aimed to isolate CTC from the blood of patients suffering from epithelial and haematological tumors, to develop protocols for the ex-vivo expansion of these cells and to perform a complete molecular characterization in order to clearly define the mutational and transcriptional profiles. This project aims to develop diagnostic kits as well as to identify new molecular targets that may be the target of innovative therapeutic tools for the treatment of these tumors. The project also includes a phase which will identify molecules with anti-tumor activity against CTC, by screening libraries of compounds, substances and natural peptides including natural products arising from innovative sources such as marine microalgae.
Descriptions of activities
The project activities are divided into six objectives (OR).
O.R. 1 Development of nanotechnological devices for the isolation of circulating tumor cells (CTC)
OR2 Molecular characterization of CTC: identification of molecular targets
OR3 Molecular characterization of CTC: development of advanced diagnostic tools
OR4 Development of mouse models for the characterization of CTC
O.R. 5 Development of new technological procedures for targeting and CTC for the optimization of
delivery of drugs and / or molecules with antitumor activity
O.R. 6 Molecular Targeting of CTC
The Zoological Station is involved in the following activities:
Activity 6.2 Identification of molecules of natural and / or synthetic origin with possible biological activity on CTC cells.
The main objective of this activity is to provide extracts and fractions from marine cultivable microalgae with anti-tumor properties, and to isolate and characterize these compounds through a screening system based on biological assays of the fractions obtained from crude extracts. Finally we plan to optimize the production of bioactive compounds of interest through cultivation on a large scale of the producer microorganism(s).
Expected Results
Identification of at least one small molecule with antitumor activity on CTC cells.
Time chart

Tab 1 Costs
| Costs | Value | Duration |
|---|---|---|
| Total personnel costa | 200.000 | 36,00 |
| Total sub-contracting costs | 200.000 | 36,00 |
| Other costs | 160.000 | 36,00 |
| Equipments | 40.000 | 36,00 |
| General Costs | 100.000 | 36,00 |
| Total | 700.000 |
Tab 2 Personnel SZN
| MM/person | ||
|---|---|---|
| Ianora Adrianna | Senior researcher | 4 |
| Romano Giovanna | Researcher | 5,5 |
| Esposito Francesco | Technologist | 4 |
| Palumbo Flora | CTER | 7,6 |
| Perna Massimo | CTER | 7,6 |
| Biffali Elio | Technologist | 3 |
| Marino RIta | Technologist | 3 |
| Pannone Raimondo | CTER | 4 |
| Mauriello Elvira | CTER | 3 |
PON01_00117 - Novartis
PON01_00117 Antigens and Adjuvants for Vaccines and Immunotherapy
Leader
Novartis Vaccines & Diagnostics Ltd.
Duration
36 months
Keywords
Biotechnology, human health, microalgae
ERC sectors
LS7_3, LS6_11, LS7_9
Summary of the project
The therapeutic benefits of a stimulation of the innate immune system are well documented. Many substances of natural origin, including some lipidic or glycolipidic molecules are able to stimulate various cellular components of the innate immune system. Therefore metabolites of low molecular weight or lipid components of extracts of marine eukaryotic microorganisms may have activating properties of the innate immune cells and act as adjuvants or immune-modulators.
The innovative approach proposed in this project is based on a public-private partnership and belongs mainly to Areas of Convergence (Sicily, Calabria and Campania).
The project aims to achieve three important goals: 1. The development of innovative vaccines for bacterial and viral infections 2. The development of new molecules with adjuvant action and the study of the mechanism of action of those already known 3. The development of more effective and safe new viral vectors for the development of new vaccines.
The role of the Zoological Station in the project is to identify and cultivate species of marine microalgae that show potential antigen and adjuvant activities.
Descriptinos of the activity
The project involves 3 objectives:
OR 1. Identification of antigenic candidates for the development of potential vaccines against Streptococcus pneumoniae, Clostridium difficile, Escherichia coli pathogens, HCMV
OR 2. Identification of new adjuvants and analysis of the mechanism of action of known adjuvants
OR 3. Identification and validation of new Adenovirus and MVA vectors and Virus Like Particles for use as potential vaccines against infectious diseases (HCMV, Malaria, Influenza, RSV, etc.)
The Zoological Station participates in activity 2.1
Activity 2.1. Identification of novel natural compounds with immune-modulatory and adjuvant activity from marine microalgae
and in particular in:
RI 2.1.1. Preparation of extracts and fractions from microalgae, isolation and identification of substances with immune-regulatory properties (Stazione Zoologica).
Extracts of marine organisms have already been shown to contain immune-regulatory substances (eg alpha galoctoside ceramide that can stimulate NKT cells) and lipids other than those present in humans. This project aims not only to identify new molecules able to interact with the immune system, but also to identify compounds for therapeutic formulations, for example compounds as adjuvants for vaccines. To this end, the group of dr. Ianora is preparing pellets of microalgae for the isolation and identification of substances with potentially immune-regulatory activity. The fractions are assayed by industrial partners (Novartis) for their ability to activate various cells and receptors of the innate immune system. The study will be performed in collaboration with IBB-CNR (SZN sub-contractor) who will characterize the various active components through the use of NMR techniques.
Time chart
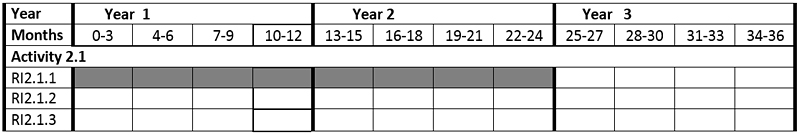
Expected results
To obtain at least one new natural compound from marine microalgae with immuno-stimulatory or adjuvant activity.
Tab 1 Costs
| Costs | Value | Total duration (months) |
|---|---|---|
| Total personnel costs | 435.000 | 24 |
| Total sub-contracting costs | 140.000 | 24 |
| Other costs | 318.500 | 24 |
| General costs | 217.500 |
Tab 2 Personnel SZN
| Total MM/person | ||
|---|---|---|
| Ianora Adrianna | Senior researcher | 8 |
| Romano Giovanna | Researcher | 8 |
| Esposito Francesco | Technologist | 10 |
| Palumbo Flora | CTER | 10 |
| Perna Massimo | CTER | 10 |
| Cter 2 temp determinato | CTER | 22 |
PON 02093 - Sanofi
PON01_02093 Study of new technologies and technological platforms for the improvement of production processes of active pharmaceutical ingredients of industrial interest and search for new bioactive molecules from natural sources
Leader
Sanofi-Aventis S.p.A.
Duration
36 months
Keywords
natural medicines, chemotherapy, microalgae
ERC sectors
LS7_3, LS9_9
Summary of the project
The overall objective of the project is the study and application of advanced and innovative technologies for the improvement of productive processes of pharmaceutical industry and the search for new molecules with potential pharmacological activity in the anti-infective, anti- cancer and anti-inflammatory field.
The first research line of the project will study the most innovative aspects of technologies of microbiology and genetics of producer strains.
The second research line will study the possibility to identify new products as candidates of potential pharmaceutical interest in the fields of anti-infectives and more generally in cancer and chronic-degenerative diseases connected with ageing, with particular attention to inflammation role. These activities will be focused on the search for new substances of pharmacological interest by screening extracts from microorganisms and / or aquatic organisms and on the characterization of their beneficial and anti-infection properties.
The project will allow the maintenance of a high scientific and technological concentration of great innovative potential characterized by an organic collaboration between industrial and academic researchers.
Description of activities
The project is divided into two lines, each divided into four development objectives (OR) according to the following scheme:
OR 1.1 - Genetic Improvement / genomic technologies
OR 1.2 - Genetic Improvement / Strain improvement
OR 1.3 - Physiology of fermentation
OR 1.4 - Extraction / Purification
OR 2.1 - New methods for the search of bioactive molecules by microorganisms
OR 2.2 - Summary of chemical derivatives of mature products
OR 2.3 - Identification of target and natural compounds relevant to cancer diseases, and chronic degenerative diseases associated with aging
OR 2.4 - Screening and characterization of extracts from marine organisms
The Zoological Station will be involved in the following activities:
OR 2.4 - Screening and characterization of extracts from marine organisms
The purpose of this Objective is the identification of new active principles with antimicrobial and / or antitumor and / or protective activity against neurodegeneration and / or aging. To achieve this goal the Zoological Station will perform the following activities:
ARI 2.4.1 - Identification of bodies from which to extract the active ingredients and their collection
Will be identified already known that microalgae can be a source of active ingredients with antimicrobial activity, antitumor, antineurodegenerativa, anti-aging. The selection will be made on the basis of their environmental activities.
ARI 2.4.2 - Extraction and sample preparation
Each species of microalgae will be cultivated to SZN so as to obtain a sufficient biomass for the extraction of small molecules by the Institute of CNR of Biomolecular Chemistry (ICB-CNR) as a third party custodial .. Once you have identified the active fractions from the other partners will identify molecule chemistry by the ICB and production in the SZN algae that produce these molecules.
Expected Results
Identification of at least one product that has the characteristics to be evaluated as a new "lead candidate" (New Product).
Time charts
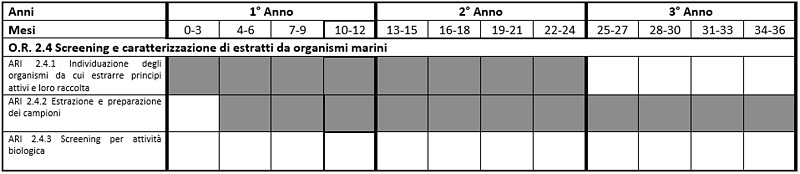
Table 1 Costs
| Costi | Valore | Durata mesi |
|---|---|---|
| Spese di personale totale | 360.000 | 36 |
| Costi consulenza tot | 225.000 | 36 |
| Altri costi esercizio totali | 160.000 | 36 |
| Spese generali | 180.000 | 36 |
Table 2 Personnel SZN
| Nominativo | Qualifica | Monte ore tot per persona | Durata mesi totale |
|---|---|---|---|
| Ianora Adrianna | Senior Researcher | 5,5 | 36 |
| Romano Giovanna | Researcher | 8 | 36 |
| Esposito Francesco | Technologist | 8 | 36 |
| Palumbo Flora | Cter | 8 | 36 |
| Perna Massimo | Cter | 8 | 36 |
| Tecn 1 temp det | Technologist | 22 | 24 |
| Cter 1 tempo det | Cter | 22 | 24 |
| Totale ore | 82 | 36 |
EMSO MedIT

Project financed in the framework of PON R&C 2007-2013 - PAC Enhancement of public research infrastructures
Coordinator Raffaella Casotti
The project concerns the enhancement of research infrastructures for marine environment located in the Convergence Regions Sicily, Campania and Puglia, where the sea is a primary opportunity of development. The project, called EMSO‐MedIT is the Italian contribution to the consolidation in the above mentioned regions of the European research infrastructure EMSO, which, within the context
of EMSO‐MedIT, is in synergy with the other ESFRI coordinated by Italy (KM3NeT and EMBRC) and the Italian initiative for marine research RITMARE.
The actions foreseen will be carried out according to the following objectives:
1) enhancement of marine infrastructures and of scientific and technological facilities to strengthenand expand the network for multidisciplinary monitoring of coastal, deep and the water column
marine environment;
2) networking of all existing and enhanced infrastructures for the transmission in real‐time/near‐realtimeintegrating the data from fixed and relocatable observing systems;
3) establishment of a mobile system of intervention to be used for monitoring campaigns at sites ofstrategic interest or in the case of environmental emergencies.
The network of monitoring infrastructures will be further enhanced through the creation of anexchange information system that will enable the sharing of the large amounts of data, providing access to a large community of Italian and foreign users.
Our role: We are partners of the project and in charge of the WP2 "Strengthening of Campania", together with INGV for the Gulf of Pozzuoli. The expansion includes the acquisition of different oceanographic instrumentation, including a WaveGlider, an ROV, and various sensors, but mainly two buoys type elastic beacon to locate in the Gulf of Naples and Gulf of Pozzuoli for the real-time monitoring and data transmission to the control center of physical and biological environmental data.
Partners: National Institute of Geophysics and Volcanology (INGV), Anton Dohrn Zoological Station (SZN), National Institute of Nuclear Physics (INFN), the National Research Center (CNR), National Institute for Environmental Protection and Research (ISPRA)
IRMA
IRMA: Implementation and Remote Connection for Real Time Moniotoring of Marine Microorganisms
Funded by the Italian Ministry of University and Education (MIUR)
Scientific Coordinator Raffaella Casotti
This project proposes to use a prototype for the automatic, continuous staining of bacteria for the in situ high frequency monitoring (several times a day, up to every 30 min). This instrument will complement the tools already available to SZN for the monitoring of photosynthetic microorganisms, allowing the definition in close temporal and spatial scale of the microbial compartment. We will study the feasibility of use on sampling boats, even without any technical supervision and, in the long term, on buoys. The data produced will provide useful information on the microbial dynamics at very small time scale in several coastal sites. The final product will be a demonstrator of multiparameter monitoring station that transmits environmental data from different sources in real time, operated remotely, helping to build an early warning system for environmental risk
Our role in the project is to coordinate actions and to test, calibrate and validate the prototype
Partners: SZN, CytoBuoy bv (Holland, George Dubelaar), Eawag (Switzerland, Frederick Hammes), CNRS- MIO (France, Gérald Grégori, Melilotus Thyssen, Michel Denis), INGV (Italian, Giovanni Iannaccone, Sergio Guardato)
S&T MED
Financed by the ENPI CBC Mediterranean Sea Basin 2007/2013 Program of the European Union
Scientific Responsible Raffaella Casotti
The project aims to provide the necessary technical and organizational support, especially to small and medium-sized enterprises (existing and potential) that operate in the food sector and in other traditional sectors of tourism, to promote coordinated actions aimed at increasing the share revenue from tourism which benefits the economies of the countries of the Mediterranean. The main result of the project is the creation of new business activities related to sustainable tourism in four coastal areas with significant natural and cultural resources and the strengthening of alliances with companies through the adoption of an approach to public / private management, quality standards and incentive systems that are coupled objectives of economic development with the protection and enhancement of natural / cultural heritage.
In addition, the project will create an international network of sustainable coastal tourism destinations in the Mediterranean Sea basin as a place for the development of common methodologies during project implementation and in the long run such as monitoring and promotional platform owned by local users, national and scientific and open to new subjects, that could spur further development of sustainable coastal tourism on the Mediterranean route.
We are partners in the project and our role is to create the environmental monitoring system of the three sites in order to a) provide real-time data on environmental conditions b) raise awareness among visitors to environmental issues, spreading the sceintific culture through actions such as "citizen science" c) train tourism operators so that they are aware of the environmental value of their sites and participate in their development and conservation
Partners: the Ministry of Cultural Heritage and Activities (coordinator), the Sinis Peninsula (Sardinia), the Zoological Station Anton Dohrm, the city of Mahdia (Tunisia) and the National Institute of Marine Sciences and Technology of Tunisia , the Al-Balqa Applied University and the Marine Protected Area of Aqaba (Jordan)
DiaEdit
Summary
The DiaEdit project, Development of genetic tools for the establishment of routine genome editing in the marine diatom Phaeodactylum tricornutum, is part of the initiative “Increasing the Potential of Marine Microeukaryotes as Experimental Model Systems through the Development of Genetic Tools” promoted by the Gordon and Betty Moore Foundation.
The recent development of genetic tools for targeted genome editing of diatoms constitutes a great opportunity for the characterization of molecular processes in these ecologically important algae. Genome editing technologies in diatoms, however, are still in their infancy regarding their routine application. Targeted mutagenesis in diatoms is challenging because of their mostly diploid state and the current lack of efficient homologous recombination.
In this project we propose to enlarge knowledge and tools for genome editing in the molecular model species Phaeodactylum tricornutum, an essential requirement to transfer these technologies to other diatoms. We plan to develop and/or validate three different approaches for genome editing: a TALEN-based approach, the utilization of CRISPR/Cas9 and a viral integrase system.
What we do
SZN is involved in Task 4 "Control of nuclease expression", aimed at the improvement of the specificity of action and expression of the nuclease used to modify the genome. This will be done mainly by identifying promoters that can allow fine control of the nuclease expression. NGS (next generation sequencing) will be used to assess the level of specificity of the chosen system by re-sequencing engineered clones.
Partners
SZN; Universitè Pierre et Marie Curie Paris, France; University of Konstanz, Germany; Norwegian University of Science and Technology, Norway; Tel Aviv University, Israel; Biological Systems and Biochemical Engineering Laboratory INSA/CNRS, France.
Research Area
Functional Genomics, Marine Biotechnology
Project Lifetime
October 2015 - September 2017
SZN Role
Partner
SZN Principal Investigator
Project Leader
Angela Falciatore, UPMC
Funding Institution
The Marine Microbiology Initiative funded by the Gordon and Betty Moore Foundation (USA).
Dedicated website
Under construction
Personnel involved
Mariella Ferrante, Principal investigator
Monia Russo, Senior Post-doc
EMBRIC
Summary
EMBRIC, European Marine Biological Research Infrastructure Cluster to promote the Blue Bioeconomy, is a large project with the overarching objective of building interconnectivity along three dimensions: science, industry, and regional RDI policies. The expected endpoint is the formation of a perennial cluster of research institutes (RIs), which will foster innovation in marine biotechnologies. To prepare this sustainable cluster, EMBRIC focuses on two specific sectors of marine biotechnology, namely (i) discovery and development of marine natural products, and (ii) marker-assisted selection in aquaculture.
SZN is involved in WP7 and WP10.
The objective of WP7 is to demonstrate that linking complementary expertise in biology, analytical chemistry and genetic engineering can provide the blue biotechnology industry with high-performance strains from across the diversity of microalgae. This will involve:
1) Proof of concept that strains from across microalgal diversity constitute a rich resource of natural products for commercial exploitation.
2) Proof of concept that microalgal strains can be genetically engineered to improve their performance capabilities for commercial exploitation.
3) Proof of concept that selective breeding in microalgae in combination with genotype screening can produce strains with improved performance in commercial applications.
The objective of WP10 is to demonstrate that:
1) The external scientific user community is interested in using EMBRIC, i.e., infrastructure workflows across multiple RI-partners.
2) RIs within EMBRIC do provide integrated, interoperable transnational access.
3) Translational access to EMBRIC combined with interdisciplinary collaboration with in-house researchers can initiate Key Enabling Technologies.
4) Translational access aids external users with the maturation of their ideas for Technology Transfer.
What we do
Within WP7, the SZN will contribute to the identification of bioactive compounds from microalgal strains and to the generation of genetically engineered diatom strains.
Within WP10, the SZN will manage the translational access, coordinating the scientific, technical and logistic access to the different RIs involved.
Partners
27 partners from seven European countries plus one Israeli institution are involved in this project.
Research Area
Marine Biotechnology
Project Lifetime
June 2015 – May 2018
SZN Role
Partner
Principal Investigator
Wiebe Kooistra, WP10 and Steering Committee member for SZN
Mariella Ferrante, WP7
Project Leader
Bernard Kloareg, France
Funding Institution
European Commission, under the H2020-INFRADEV-4 call
Dedicated website
Personnel involved
Mariella Ferrante, Researcher
Wiebe Kooistra, Senior Researcher
Adrianna Ianora, Senior Researcher
Marina Montresor, Senior Researcher