| Descrizione |
Le unità IRM e MAA possiedono una pluriennale esperienza nel campo del monitoraggio ambientale maturata in seguito alla partecipazione a numerose commesse esterne e progetti scientifici. L’attività consiste nella raccolta, analisi ed elaborazione dati finalizzata al trasferimento di relazioni tecnico-scientifiche ad istituzioni locali e nazionali o a soggetti privati. Vengono effettuati:
|
| Servizi erogati |
|
| Attrezzature utilizzate |
L’attività di campionamento e raccolta dati è svolta prevalentemente a bordo della M/N Vettoria o su altre navi oceanografiche. In alternativa, viene allestito un laboratorio mobile autosufficiente per il primo trattamento dei campioni in assenza di strutture di appoggio idonee. Le attrezzature utilizzate comprendono, per il lavooro di campagna:
E, in laboratorio: |
| Contatti |
Fabio Conversano Tel. + 39 081 5833357 e-mail: fabio.conversano(at)szn.it |
Chair
Prof. Axel Meyer
Prof. Vincenzo Saggiomo
Dott. Graziano Fiorito
Alumni
Maria Grazia Mazzocchi
Marina Montresor
Maurizio Ribera d'Alcalà
Scipione Beatrice
Research Fellow
|
Aguzzi Jacopo Andrews Paul Bartoli Marco Boni Raffaele Caianiello Silvia Calò Antonio Cerrano Carlo Christiaen Lionel Crispi Stefania Donadio Carlo Faggio Caterina Feuda Roberto Fiorentino Fabio Giordano Daniela |
Iandolo Donata Iovino Nicola Mojetta Angelo Renato Rizzo Lucia Scipione Maria Beatrice Signorini Silvia Giorgia Swartz Zachary S. Taviani Marco Verde Cinzia
|
Updated on August, 2025
Unità Infrastrutture di Ricerca a Mare
| Descrizione |
Acquisizione di dati idrografici, raccolta e pretrattamento di campioni di acqua da destinare alle successive analisi delle principali variabili fisiche, chimiche e biologiche. L’attività è svolta prevalentemente a bordo della M/N Vettoria o su altre navi oceanografiche. Inoltre, in assenza di strutture di appoggio idonee, è possibile allestire un laboratorio mobile completamente autosufficiente che consente di effettuare il primo trattamento dei campioni. |
| Servizi erogati |
|
| Attrezzature utilizzate |
Imbarcazioni oceanografiche equipaggiate Sonda multiparametrica e campionatore |
| Contatti |
Fabio Conversano Tel. + 39 081 5833357 e-mail: fabio.conversano(at)szn.it |
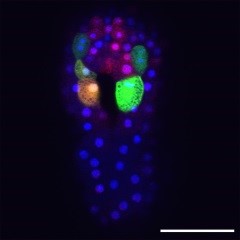
| Descrizione | L’Unità fornisce ai ricercatori tecniche aggiornate di microscopia ottica, elettronica, live cell imaging, microscopia confocale, microscopia 3D e 4D per la comprensione della struttura tridimensionale e i componenti subcellulari. |
| Servizi erogati |
- Assistenza tecnica agli utenti dei microscopi confocali - Assistenza tecnica agli utenti del microscopio a epifluorescenza corredato di sistemi di acquisizione di immagini - Consulenza per l’applicazione delle tecniche di microscopia ottica e confocale - Consulenza sull’analisi digitale delle immagini ottenute dalle osservazioni al microscopio - Formazione sull’uso delle attrezzature. |
| Attrezzature utilizzate |
- Microscopio Confocale Zeiss LSM 510 - Microscopio confocale Zeiss LSM 700 |
| Contatti |
Elio Biffali Tel. +39 081 5833 298 e-mail: elio.biffali(at)szn.it |